Structure-function characterization of membrane binding proteins
 Structure-function relations of three members of the dynamin family : The study defined the GTPase, self-assembly, and membrane binding activities of each protein, aiming to better understand the different mechanisms of action. We found that different proteins of the dynamin family form distinct highly ordered protein-membrane complexes that are customized to fit their diverse biological functions: Dynamin facilitates fission of vesicles (A&B), MxA forms stable membrane-association tubes as storage form in the pre infectious stage (C), Mgm mediates fusion of the inner mitochondrial membrane and maintenance of cristae structures (D).
Structure-function relations of three members of the dynamin family : The study defined the GTPase, self-assembly, and membrane binding activities of each protein, aiming to better understand the different mechanisms of action. We found that different proteins of the dynamin family form distinct highly ordered protein-membrane complexes that are customized to fit their diverse biological functions: Dynamin facilitates fission of vesicles (A&B), MxA forms stable membrane-association tubes as storage form in the pre infectious stage (C), Mgm mediates fusion of the inner mitochondrial membrane and maintenance of cristae structures (D).
Membrane remodeling by F-BAR domain proteins
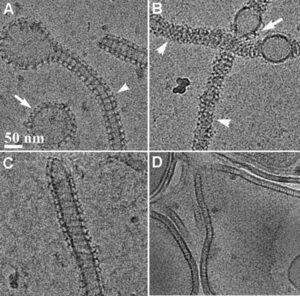
Membrane remodeling is critical for cellular processes such as cargo trafficking, signaling, cell motility, and organelle biogenesis, and it requires the concerted action of scores of proteins that bind and actively shape cellular membranes. F-BAR domain proteins regulate and sense membrane curvature by interacting with negatively charged phospholipids and assembling into higher-order scaffolds. However, regulatory mechanisms controlling these interactions are poorly understood. Here, we show that Drosophila Nervous Wreck (Nwk) is autoregulated by a C-terminal SH3 domain module that interacts directly with its F-BAR domain. Surprisingly, this autoregulation does not mediate a simple “on-off” switch for membrane remodeling. Instead, the isolated Nwk F-BAR domain efficiently assembles into higher-order structures and deforms membranes only within a limited range of negative membrane charge, and autoregulation elevates this range. Thus, autoregulation could either reduce membrane binding or promote higher-order assembly, depending on local cellular membrane composition. Our findings uncover an unexpected mechanism by which lipid composition directs membrane remodeling. The Nwk F-BAR self-assembles into zigzags, distinct from canonical F-BAR proteins, and thus induces membrane scallops and ridges rather than membrane tubules.
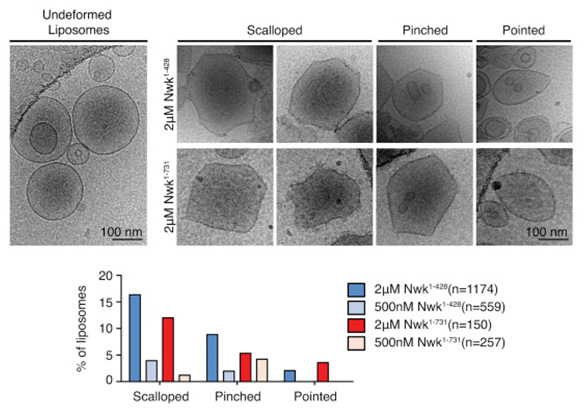
Figure: Cryo-EM of control and Nwk deformed liposomes. Both purified Nwk1–428 (F-BAR) and Nwk1–731 (full length) induce membrane scalloping, pointing, and pinching of 10% PI(4,5)P2 liposomes. Scale bar, 100 nm. Bar graph summarizes vesicle morphology after 30-min incubation of Nwk1–428 or Nwk1–731 (2 μM and 500 nM) with 0.3 mM [DOPC:DOPE:DOPS:PI(4,5)P2] = 70:15:5:10 liposomes. n represents the number of liposomes examined.